Page 2 of 2
Re: Decode of trajectory
Posted: Mon Jun 30, 2014 3:16 pm
by bruce
Do you have a backup of the original trajectory data (created by the FahCore) or have the original files been deleted? We need to capture examples like that one.
It looks like that's a corner-case (in more ways than one

). I'll bet that the bond is correct, but the filter missed it. It probably shows exactly how the piece on the right should be joined to the piece on the left through a pair of external box faces.
If you relocate the piece on the right in the proper direction by the dimension of the box I'll bet it creates the proper length bond and stitches those two pieces back together although now the protein is no longer completely inside the box.
Re: Decode of trajectory
Posted: Mon Jun 30, 2014 8:56 pm
by ChristianVirtual
Actually it is still crunching:
Project 8580 (Run 1, Clone 7, Gen 474)
What files you need exactly ?
Re: Decode of trajectory
Posted: Mon Jun 30, 2014 9:20 pm
by bruce
Pause the WU. Make a backup copy, along with the .db that says it's there. Resume work and let it finish.
Are you and Jesse coordinating your development efforts?
Re: Decode of trajectory
Posted: Mon Jun 30, 2014 9:45 pm
by ChristianVirtual
PM send with link to G-Drive, 20MB tar file.
Re: Decode of trajectory
Posted: Tue Jul 01, 2014 1:50 am
by Jesse_V
Thanks. The protein split glitch is a long-standing bug, and it'd be nice to track it down and fix it.
Re: Decode of trajectory
Posted: Tue Jul 01, 2014 3:26 am
by ChristianVirtual
Have two more folding:
Project: 8574 (Run 1, Clone 6, Gen 473) and
Project: 8574 (Run 1, Clone 8, Gen 500)
Will pack them tonight when coming home.
Re: Decode of trajectory
Posted: Tue Jul 01, 2014 5:02 am
by bruce
Are you saying that these proteins also split into separate segments?
AND/OR
have bonds that stretch almost from one side of the box to the opposite side?
Re: Decode of trajectory
Posted: Tue Jul 01, 2014 10:29 am
by ChristianVirtual
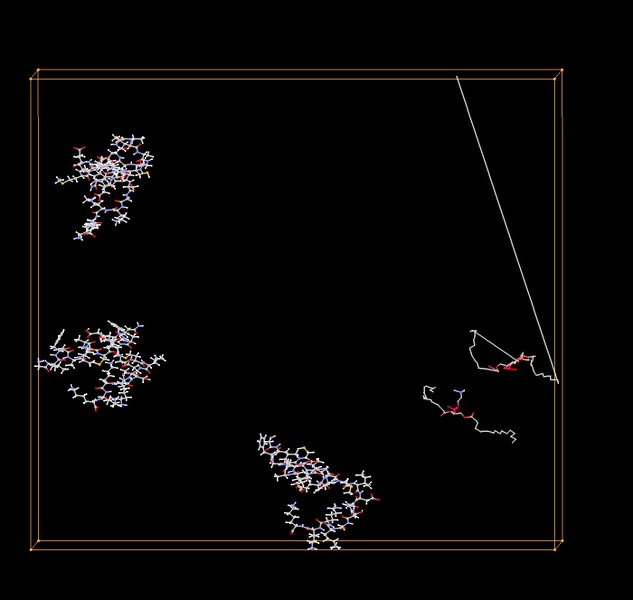
Here the one with gen 500
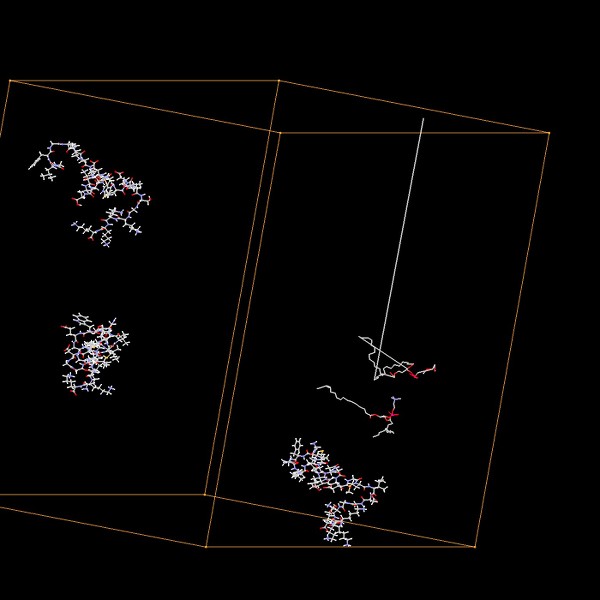
Different angle, same bond touch two borders
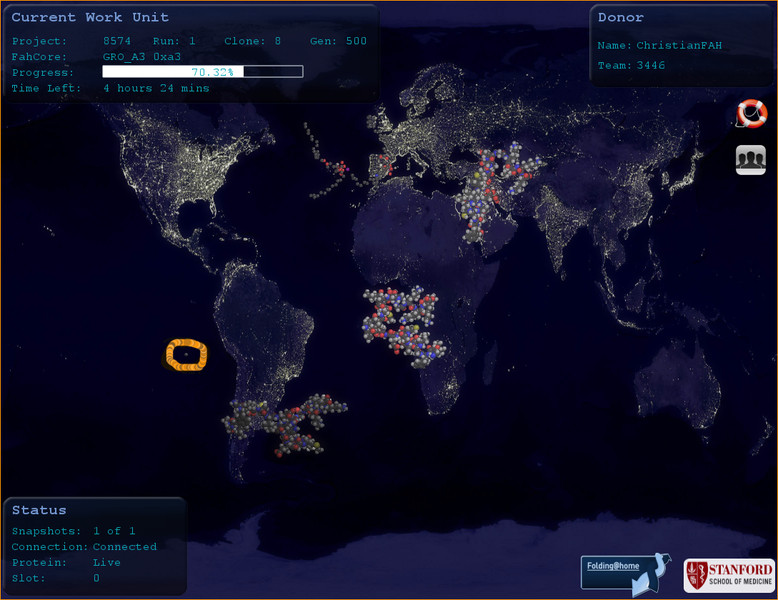
to cross check: also the official viewer has the phantom atom; circled here in orange; west coast of Latam
Re: Decode of trajectory
Posted: Tue Jul 01, 2014 3:25 pm
by bruce
Great examples. That should help a lot in fixing the bug.
Re: Decode of trajectory
Posted: Wed Aug 13, 2014 9:58 pm
by ChristianVirtual
Here comes another interessting one: seems overlapping squares ? Or missing bond.
When I rotate the protein it seems the little chain is partially embedded by the bigger chain (part of chain)
Project 6381 (Run 24, Clone 0, Gen 20)
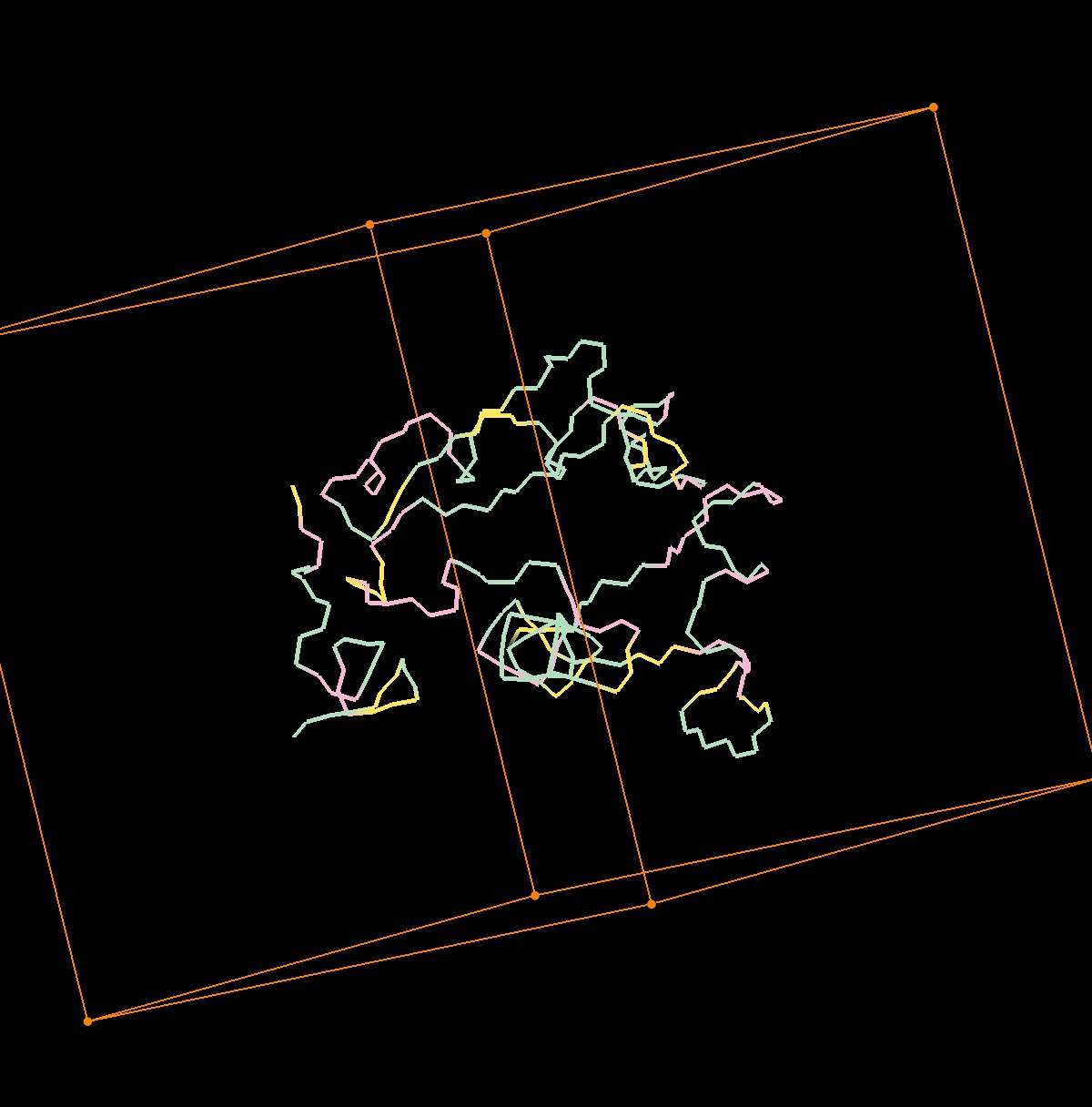 http://imageshack.com/a/img661/1545/PXlUcp.jpg
http://imageshack.com/a/img661/1545/PXlUcp.jpg
Different angle
http://imageshack.com/a/img674/4948/QyWQmN.jpg
Re: Decode of trajectory
Posted: Wed Aug 13, 2014 10:17 pm
by bruce
ChristianVirtual wrote:Here comes another interessting one: seems overlapping squares ? Or missing bond.
Not "or" ... I expect it's "and"
If you look parallel to a side of the box, the shorter piece probably has one end very, very close to that face of the box and the longer piece probably attaches to the opposite face of the box. Find the atom on each chain that attach them to a face and manually add a bond between them.
If you know how to read the data created by the FahCore, I'll bet that bond is present in the original data created by the FahCore but hidden by FAHClient during the conversion to the API data. That's a bug that needs to be documented.
Re: Decode of trajectory
Posted: Fri Aug 15, 2014 10:39 am
by ChristianVirtual
Installed the GROMACS 4.6.5 tools via Ubuntu-repository.
With those tools is rather simple to read the files from the working directory. The first learning: there can be more then one protein chain in the WU and sometimes something else (and not only water).
Like in P7505 there is a compound included called POPC (on the left); that one has quite a number of carbon atoms (comes at no surprise: 42

) It looked wired the first times I saw it but it start to make sense.
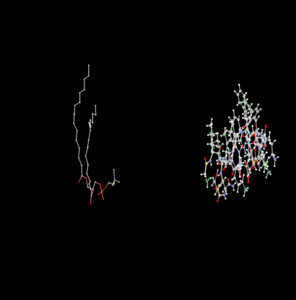
One problem is that the trajectory provided by the API don't contain those additional information like number of protein chains, the identification of the side chains/residue or additional relevant molecules. GROMAS file has it all. Passthrough would be great as it would allow much simpler and diverse visualisation.
Anyway: I'm getting used to the files and continue my journey into the "boxes" ...
The learning continues ...
Re: Decode of trajectory
Posted: Sun Aug 24, 2014 2:11 pm
by ChristianVirtual
One question: are the coordinates in trajectory in Angstrom ?
Re: Decode of trajectory
Posted: Sun Aug 24, 2014 3:15 pm
by Jesse_V
ChristianVirtual wrote:One question: are the coordinates in trajectory in Angstrom ?
I believe so.
Re: Decode of trajectory
Posted: Wed Nov 12, 2014 12:57 pm
by ChristianVirtual
Now here comes a totally messy one, lots of long bonds across the molecule, not only from side to side. Maybe still, as we dont know the size of the (repeating) box. don't open the video on mobile, bigger one
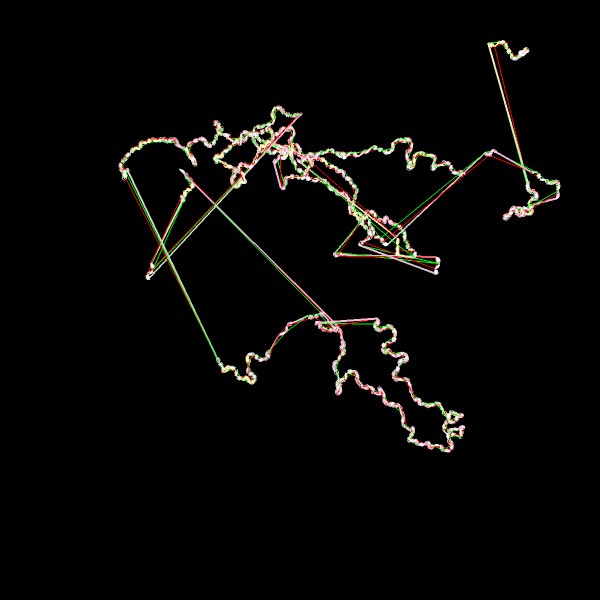 http://youtu.be/I51xmgYohuA
http://youtu.be/I51xmgYohuA
And in the second half the frame animation is swited on. Interesting that the protein just move along the y-axis.
 ). I'll bet that the bond is correct, but the filter missed it. It probably shows exactly how the piece on the right should be joined to the piece on the left through a pair of external box faces.
). I'll bet that the bond is correct, but the filter missed it. It probably shows exactly how the piece on the right should be joined to the piece on the left through a pair of external box faces.




